
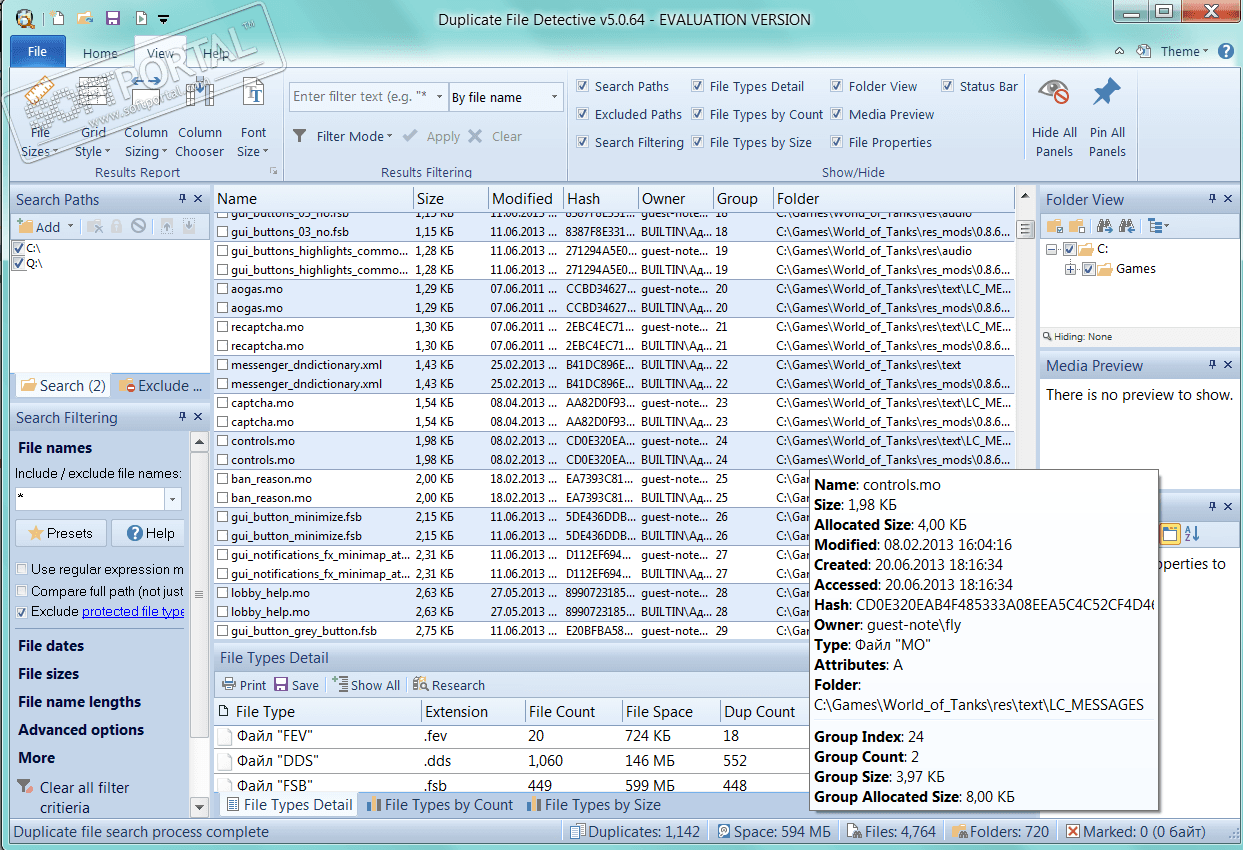
(that is, all reads with the same queryname are together in the input file). MarkDuplicatesSpark is optimized to run on queryname-grouped alignments Multithreading on the Biowulf cluster, it is necessary to add -spark-master local to the base command line. Quality and there aren’t Spark versions of each GATK tool. However, not all Spark based tools are considered production Used for parallelism in GATK 4 and can speed up the process relative to the Here we use MarkDuplicatesSpark instead of MarkDuplicates. Reads are tagged but not removed from the alignment. ( MarkDuplicates) to mark all the other reads from a set of duplicates withĪ tag. To take only one representative read, GATK uses a Picard tool Measurement bias and reduce error, only one best copy should be kept. There are two types of duplicates: PCR duplicatesĪnd Sequencing (various optical confusions) duplicates.
1.4.2 Alternative B: BAM output without coordinate sorting.1.4.1 Alternative A: producing sorted BAM output.

1.3.4 Alternative B: Queryname-grouped (unsorted) BAM.1.3.3 Alternative A: Sorting by coordinate.


 0 kommentar(er)
0 kommentar(er)
